
Luciana Negrão Frota de Almeida1; Rachel Melilo Carolino2; Diogo Cazelli Sperandio3; Márcio Bittar Nehemy4; LA De Marco5
DOI: 10.1590/S0004-27492009000400027
ABSTRACT
Age-related macular degeneration (AMD) is the most frequent cause of irreversible blindness in the elderly in developed countries. Although the etiology of AMD remains largely unknown, numerous studies have suggested that both genes and environmental risk factors significantly influence the risk of developing AMD. Recently, single nucleotide polymorphisms, DNA sequence variations found within the complement factor H (CFH) gene, have been found to be strongly associated with the development of AMD. Several other genes have had at least one positive association finding and deserve further exploration. The purpose of this review is to provide an extensive report of the current data of AMD genetics and the contribution of this knowledge helps to the better understanding of its pathophysiology.
Keywords: Macular degeneration; Age effect; Complement factor H; Base sequence; Polymorphism, single nucleotide; Inflammation
RESUMO
A degeneração macular relacionada à idade (DMRI) é a causa mais frequente de cegueira irreversível em idosos em países desenvolvidos. Apesar da etiologia da DMRI ainda permanecer desconhecida, numerosos estudos tem sugerido que tanto fatores genéticos quanto ambientais influenciam significativamente no risco do desenvolvimento da doença. Recentemente, polimorfismos de base única, variações na sequência de DNA encontradas no gene fator H do complemento (CFH), tem sido fortemente associado com o desenvolvimento da DMRI. Muitos outros genes tiveram ao menos um resultado positivo para esta associação e merecem estudos posteriores. O objetivo dessa revisão é proporcionar descrição atual dos dados publicados.
Descritores: Degeneração macular; Fator idade; Fator do complemento H; Sequência de bases; Polimorfismo de um único nucleotídeo; Inflamação
ATUALIZAÇÃO CONTINUADA
The role of molecular genetic factors in age-related macular degeneration
A importância dos fatores genéticos na degeneração macular relacionada à idade
Luciana Negrão Frota de AlmeidaI; Rachel Melilo CarolinoII; Diogo Cazelli SperandioIII; Márcio Bittar NehemyIV; De Marco LAV
IPós-graduanda (Doutorado) em Cirurgia e Oftalmologia da Universidade Federal de Minas Gerais - UFMG - Belo Horizonte (MG) - Brazil
IIAcadêmica, Aluna de Iniciação Científica do Laboratório de Farmacologia Bioquímica e Molecular da UFMG - Belo Horizonte (MG) - Brazil
IIIResidente em Oftalmologia do Hospital São Geraldo da UFMG - Belo Horizonte (MG) - Brazil
IVProfessor Adjunto e Chefe do Serviço de Retina e Vítreo do Hospital São Geraldo da UFMG - Belo Horizonte (MG) - Brazil
VProfessor Titular e Coordenador do Programa de Pós-graduação em Farmacologia Bioquímica e Molecular da UFMG - Belo Horizonte (MG) - Brazil
ABSTRACT
Age-related macular degeneration (AMD) is the most frequent cause of irreversible blindness in the elderly in developed countries. Although the etiology of AMD remains largely unknown, numerous studies have suggested that both genes and environmental risk factors significantly influence the risk of developing AMD. Recently, single nucleotide polymorphisms, DNA sequence variations found within the complement factor H (CFH) gene, have been found to be strongly associated with the development of AMD. Several other genes have had at least one positive association finding and deserve further exploration. The purpose of this review is to provide an extensive report of the current data of AMD genetics and the contribution of this knowledge helps to the better understanding of its pathophysiology.
Keywords: Macular degeneration/etiology; Age effect; Complement factor H/genetics; Base sequence; Polymorphism, single nucleotide; Inflammation
RESUMO
A degeneração macular relacionada à idade (DMRI) é a causa mais frequente de cegueira irreversível em idosos em países desenvolvidos. Apesar da etiologia da DMRI ainda permanecer desconhecida, numerosos estudos tem sugerido que tanto fatores genéticos quanto ambientais influenciam significativamente no risco do desenvolvimento da doença. Recentemente, polimorfismos de base única, variações na sequência de DNA encontradas no gene fator H do complemento (CFH), tem sido fortemente associado com o desenvolvimento da DMRI. Muitos outros genes tiveram ao menos um resultado positivo para esta associação e merecem estudos posteriores. O objetivo dessa revisão é proporcionar descrição atual dos dados publicados.
Descritores: Degeneração macular/etiologia; Fator idade; Fator do complemento H/genética; Sequência de bases; Polimorfismo de um único nucleotídeo; Inflamação
INTRODUCTION
Age-related macular degeneration (AMD) is a complex degenerative disorder that primarily affects the elderly(1). Disease susceptibility is influenced by multiple genetic and environmental factors(1-2). Despite of success in identifying some genetic basis of sub-groups of AMD, investigators continue to struggle to identify other genes that predispose to, or cause the majority of this disorder(3-25).
Genetically, the condition is somewhat difficult to study because of its clinical variability and late onset, and also because AMD is a complex genetic disorder. In contrast to a classic Mendelian disease, in which one copy of an altered gene (autosomal dominant inheritance) or two copies of altered genes (autosomal recessive inheritance) are necessary and sufficient to cause a disease phenotype, in complex genetic or polygenic disorders, there can be genetic heterogeneity and variants interactions from multiple genes. In addition, genetic variants may interact with behavioral and environmental factors that contribute to enhance the disease risk, challenging the classic concept of causality that has dominated genetic research of rare disorders.
However, recent studies have identified a number of chromosomal regions harboring potential AMD genes using massive genome screening and complex statistical analysis(3-5). Candidate genes are selected based on either known characteristics of a gene (e.g., mutation predisposing to a trait) or physiologic association (e.g., genes' role in modulation of complement activation). Once a gene is selected, it is amplified and then analyzed for the presence of polymorphisms (variants of the gene). The candidate gene is then compared with genes of a population of affected patients and controls to see if a true association exists.
The purpose of this review is to provide a report of the current data of AMD genetics and the contribution of this knowledge helps to understands better its pathophysiology.
Genetic overview
Before discussing the recent discoveries regarding the genes that have been associated with AMD, it is important to consider some genetic definitions and nomenclature that are not too familiar for many ophthalmologists but are extremely important to enhance the understanding of this issue.
Deoxyribonucleic acid (DNA) is a long polymer of simple units called nucleotides made of sugars and phosphate groups joined by ester bonds. Attached to each sugar is one of four types of molecules called bases - A (adenine), C (cytosine), G (guanine), and T (thymidine). It is the sequence of these four bases along the polymer that encodes information. Sequence variation could lead to a mutation, which means "change" in some disciplines and "disease-causing change" in others; or polymorphism, which means "non-disease-causing change" or "change found at a frequency of 1% or higher in the population"(26).
A polymorphism can be found at the encoding gene sequence called exons, or at the non-coding region known as introns and could occur as substitutions, deletions, duplications or insertions of a base pair(26-27). The most common sequence variation related to AMD is the substitution of one base pair at codon 1277 of CFH gene(9-14). This type of molecular alteration is known as single nucleotide polymorphism (SNP), and is designated by a ">" character, e.g., 1277T > C or T1277C denoting that at nucleotide 1277 an T is changed to a C(9-14,26). Being a non-synonymous SNP (produces a different polypeptide sequence) the change can also be designated as Y402H denoting that amino acid 402 Tyrosine (Y) is changed to a Histidine (H)(9-14,26). All SNPs are cataloged in a public SNP database with a specific identification, for example T1277C at rs1061170(9-14).
The polymorphism can occur in a splice junction site, the DNA region that marks the boundaries between introns and exons. Sequence variation at this site can result in exon skipping or alteration of splicing (the removal of introns from heteronuclear RNA and joining of exons to form the mature messenger RNA)(27). Occasionally polymorphisms can happen close to each other, organized as a block that are transmitted together in haplotypes (the combination of linked alleles for a given region of DNA on a single chromosome)(27).
Molecular genetics of AMD
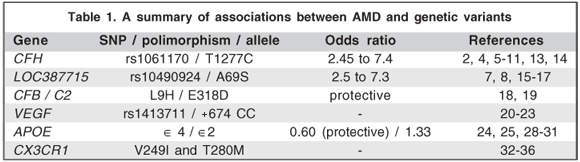
Over the past decade, researchers have begun to focus their attention on determining the genetic components of AMD (Table 1). Identification of underlying genes has been difficult, with both genomic screen (positional) and candidate gene (functional) approaches being used. Although numerous regions of interest have been identified by these genome screens, only two regions, located on chromosomes 1 and 10, have been consistently identified by the majority of studies(4-17).
CHF (complement factor H) gene
The first dramatic success of association studies for ARM was the discovery of a variant in the complement factor H (CFH) gene located in the regulation of complement (RCA) locus in chromosome 1q31.3(4,5-9). This gene encodes the CFH protein which is a major inhibitor of the complement system. It directly inhibits both classical and alternate complement pathways, therefore the absence of factor H or its low activity can lead to unchecked complement activation, serving as the inflammatory stimulus for drusen formation(2). Furthermore, environmental risk factors associated with AMD, such as smoking, are also known to decrease CFH levels in serum(2).
Recent reports have shown that a common polymorphism in exon 9 of the CFH gene (T1277C at rs1061170, or Y402H, a tyrosine to histidine change at position 402) is associated with macular soft drusen as well as an increased risk of advanced AMD, including geographic atrophy and neovascular AMD(9-13). These findings suggest that the C allele, or disease allele, contributes to almost half of all AMD cases (50 - 57.3%). The odds ratios for development of AMD in patients with the Y402H SNP ranged from 2.45 to 7.4(4-9) with higher risks seen in homozygous CC subjects(13).
Interactions with modifiable risk factors and this specific CFH polymorphism were investigated(5,8,14). The authors concluded that obesity and cigarette smoking multiplied the risks associated with these variants. It's estimated that there is a 4-fold increased risk among participants homozygous for CFH Y402H who are not obese compared with nonobese heterozygous subjects, whereas the risk is 12-fold higher among homozygous subjects who were also obese(8). Smokers of 10 pack-years or more who also have the CC genotype have approximately a 144-fold increase in disease risk compared with individuals who smoke fewer than 10 pack-years and have the CT or TT genotype(5). Although it was demonstrated that these two risk factors are independent of each other, they may work through similar mechanisms to increase the risk of neovascular AMD(14). These studies suggested that each of these factors may independently increase risk of neovascular AMD through similar mechanisms thus providing the possibility that one could potentially lessen a significant component of AMD risk by behavioral modifications(5,8,14).
In addition to Y402H, two other coding-region non-synonymous SNPs (I62V in exon 2 and D936E in exon 19) have also been described(10). These SNPs, as the Y402H, might affect the protein function through variability in expression levels, binding efficiencies, and/or other properties(10).
Several additional intronic SNPs were also related to AMD. A common variant in intron 2 splice acceptor site, IVS2-18insTT, was detected(10). Moreover, a recent publication demonstrated a previously unreported intronic SNP (rs1410996) that unexpectedly showed strong association with AMD(9).
Some authors suggested that no single polymorphism could account for the contribution of the CFH locus to disease susceptibility. Instead, multiple polymorphisms defined a set of four common haplotypes (of which two were associated with disease susceptibility and two seemed to be protective) and multiple rare haplotypes (associated with increased susceptibility in aggregate) and that noncoding CFH variants may play a role in disease susceptibility(4). It has also been suggested that not only two, but four haplotypes of the CFH gene are protective (H2, H3, H4, and H5) with the homozygous H2/H2 individual having the greatest protective effect (odds ratio, 0.27; 95% confidence interval, 0.1-0.74). Haplotype analysis shows that the most frequent at-risk haplotype is present in half of individuals with AMD, compared with 29% of controls(10).
Most of the AMD-associated CFH SNPs lie within important functional domains of the encoded protein. These domains contain binding sites for C3b, heparin, sialic acid, and C-reactive protein. Thus, these SNPs might affect CFH function through variability in expression levels, binding efficiencies, and/or other properties. For example, the exon 2 I62V variant is located in specific domain, which includes a C3b binding site, and the exon 9Y402H variant lies within a domain which binds heparin. It remains to be established how alterations in these regions might impact the function of the CFH protein. However, the absence of factor H or low activity can lead to unchecked complement activation, serving as the inflammatory stimulus for drusen formation(10).
CFH has closely related genes CFHR3, CFHR1, CFHR4, CFHR2 and CFHR5, which are arranged in tandem on chromosome 1q23. Deletions of CFHR1 and CFHR3 have been associated with decreased risk for AMD, possibly because the products of both genes, when present in the circulation have the potential to compete with CFH for C3 binding(11).
LOC387715 gene
A second putative AMD susceptibility gene, located on chromosome 10q26, LOC387715 (T allele at rs10490924, or A69S), has recently been identified(7-8). Its function is unknown, although at least one gene in close proximity to LOC387715, PLEKHA1, is known to be involved in cellular immunity. Some authors reported an odds ratio of 5 and a population attributable risk of 57% for the PLEKHA1/LOC387715 locus(15-16).
The risk allele for LOC387715 confers an OR of 2.5 (95% CI, 2.2- 2.9) and 7.3 (95% CI, 5.7-9.4) for the heterozygous and homozygous states, respectively(15). Some studies suggest that individuals with AMD possessing one or more risk alleles at LOC387715 are more likely to develop neovascular AMD compared with those with AMD who lack this variant(7). In addition, smokers with this variant have a 22-fold higher risk for advanced AMD, especially the neovascular form, compared with nonsmokers with this variant(8,17).
For patients homozygous for both risk variants, Y406H SNP and A69S SNP, the odds ratio for development of AMD was 57.58 (95% CI, 37.24-89), and they might be at greater risk for earlier onset of neovascular AMD.7 When smoking is introduced into the equation, it is estimated that CFH, LOC387715, and smoking explain 61% of the attributable risk for AMD(17). Controlling for each of the other two variables, the approximate population-adjusted risk for AMD and smoking is 20%, for AMD and A69S is 36%, and for Y402H is 43%(17).
Interestingly, a study aimed to determine whether modifiable risk factors act synergistically with LOC387715 to alter the risk of AMD suggested that regular acetyl salicylic acid use may reduce the risk conferred by homozygosity for LOC387715 A69S, and concluded that this variant appear to act synergistically with modifiable risk factors(8).
Complement factor B (CFB) and complement component 2 (C2) genes
Complement factor B (CFB) aids initiation of the alternative complement cascade, and complement component 2 (C2) activates the classical component pathway. CC2 is paralogous to CFB and resides adjacent to CFB on chromosome 6(18). In addition, they are expressed in the neural retina, retinal pigment epithelium (RPE) and choroid(19). The CFB R32Q SNP and the haplotype containing the C2 E318D/CBF L9H pair were significantly protective for AMD(18-19). After adjusting for age, Y402H, A69S and smoking, the effect of CFB R32Q remained quite strong(19) extending and refining the role of the alternative complement pathway in the pathophysiology of AMD and further strengthening the proposed model that inflammation has an important role in this ocular disease.
VEGF (vascular endothelial growth factor) gene
The vascular endothelial growth factor (VEGF) gene encodes the VEGF protein, a major player in the control of angiogenesis(1,20-22). VEGF-A contains nine exons and is alternatively spliced to produce isoforms of differing lengths, 189, 165 and 121 amino acids. The most predominant isoform in the eye is the VEGF 165 which is further spliced into an 'a' or 'b' isoforms, the difference being the presence at the C-terminus of either exon 8 or exon 9, respectively. VEGF 165a has potent angiogenic properties, whereas VEGF165b has anti-angiogenic properties(20). What controls splicing is still largely unknown but the balance between these two isoforms is vital in maintaining vascular homeostasis.
Increased levels of VEGF in surgically removed neovascular membranes from eyes with neovascular AMD have been demonstrated(23). And it has been suggested that the presence of polymorphic sequences within the promoter or 5'-untranslated region of the VEGF gene can influence levels of VEGF protein expression(21). Churchill et al.(20) showed that a specific haplotype in this region is significantly associated with AMD (OR= 18.24, 95% CI 2.25-148.25, corrected P=0.0074).
Some studies found that the CC genotype at +674 was significantly associated with the AMD group, whereas the presence of the T allele was significantly associated with the control group(20-21). These data support the hypothesis that the +674 CC genotype is a potential risk factor for AMD, and that carriage of the T allele is potentially protective(20,21).
In summary, the molecular studies associated with the functional role in vascular growth and regeneration(20-23), strongly implicates the VEGF gene in the etiology of neovascular AMD.
APOE (apolipoprotein E) gene
Apolipoprotein E (APOE) is a protein that plays a central part in plasma metabolism of lipids, in central nervous system lipid homeostasis(24) and in response to neuronal injury(25). It has been shown to be expressed in the RPE layer in mouse embryos, suggesting a role of this protein in retinal diseases(28). Three common alleles, ∈2, ∈3 and ∈4 encode the three protein isoforms E2, E3 and E4, respectively. Although some authors showed no association with AMD(29) some studies reported a reduction in the frequency of the ∈4 allele in patients with AMD, including geographic atrophy and neovascular form(30), compared to controls, suggesting its protective effect(5,23,31). In addition, ∈2 allele frequency was increased in AMD patients compared to controls, suggesting a modest increase in disease risk with this allele(31).
APOE4 lacks cystein residues at position 112 and 158, which prevents the formation of disulfide bridges with APOAII or other peptide components(24). Therefore, the inability of APOE4 to form dimers compared with APOE2 and APOE3, would favour easier transport of lipids through Bruch membrane because of a smaller size of lipid particles, allowing a better elimination and preventing of drusen formation(24-25).
CX3CR1 gene
Chemokines are small proteins that induce directed chemotaxis in nearby responsive immune cells(32). CX3C chemokine receptor 1 (CX3CR1) is expressed in microglial cells (MCs), the resident macrophages in the central neural system (CNS), and in the photoreceptor retinal layer(33-34). This receptor me-diates migration and adhesion of these cells in response to CX3C chemokine ligand 1 (CX3CL1)(32). It has been demonstrated that all retinal MCs express CX3C chemokine receptor 1 (CX3CR1)(32) and that two polimorphisms in the CX3CR1 gene, V249I and T280M, which are associated with impaired subretinal cell migration, increase the risk of AMD(32,34).
Microglial cells participate in AMD development and progression by controlling retinal MC redistribution(32) and increasing macrophage recruitment, which has previously been shown to contribute in drusen genesis and CNV development(34). However, in animal models it has been demonstrated that decreased macrophage recruitment increases choroidal neovascularization(35). These findings suggest that macrophage recruitment into the subretinal space is necessary to remove extracellular deposits and prevent the formation of drusen. These two controversy concepts could be explained by the fact that macrophages can have many different and even opposite functions depending on their activation state(35).
Decreased expression of CX3CR1 results in macrophages with a proinflammatory phenotype compared with macrophages with high levels of CX3CR1, which have an antiinflammatory phenotype(36). It is rational to consider that the accumulation in the subretinal space of resident microglia with reduced CX3CR1 function might therefore contribute to inflammation, leading to the recruitment of other inflammatory cells, which sequentially stimulate choroidal neovascularization and AMD.
ABCR gene
The presence of phenotypic similarities between Stargardt disease and AMD lead to multiple association studies between the gene responsible for the majority of cases of Stargardt disease, the ABCR gene and AMD. Some of these studies have indicated an association of AMD with certain non-synonymous SNP in ABCR gene, notably D2177N and G1961E(37). However, this association has not been confirmed by other investigators(38). To date, the ABCR variants do not seem to make a major contribution to AMD susceptibility.
Other genes
Studies that examined genes related to retinal dystrophy, such as ELOVL4 (Stargardt disease)(5), bestrophin (Best disease)(3,37), failed to firmly establish an association with AMD. In addition, some studies have suggested a role of VLDLR(3), LRP6(3) and HEMICENTIN-1(39) genes in AMD pathophysiology. However, further studies are needed to ascertain their impact on disease susceptibility(40).
CONCLUSION
Analysis of genetic susceptibility and potential interactions between variants and environmental factors (such as smoking and nutrition) seem to be essential to elucidate mechanisms of disease pathology. Association studies have provided valuable insights to demonstrate genetic differences that influence susceptibility to AMD.
A better understanding of the essential molecular mechanisms of diseases will allow us to define the relationship between the different AMD phenotypes expressed in patients. In turn, this will guide us to more accurate definitions and classification systems for AMD, leading to the development of new and effective therapies that are specific for each subtype of AMD. In addition, new preventive treatments will likely be developed using current information about the pathogenesis of the disease.
REFERENCES
1. Seddon JM, Chen CA. Epidemiology of age-related macular degeneration. In: Ryan SJ, editor. Retina. 4th ed. St. Louis: Mosby; 2006. p.1017-25.
2. Donoso LA, Kim D, Frost A, Callahan A, Hageman G. The role of inflammation in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2006; 51(2):137-52. Comment in: Surv Ophthalmol. 2006;51(5):532; author reply 532.
3. Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, et al. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006; 47(1):329-35.
4. Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9): 1049-54.
5. DeAngelis MM, Ji F, Kim IK, Adams S, Capone A Jr, Ott J, et al. Cigarette smoking, CFH, APOE, ELOVL4, and risk of neovascular age-related macular degeneration. Arch Ophthalmol. 2007;125(1):49-54.
6. Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421-4.
7. Shuler RK Jr, Hauser MA, Caldwell J, Gallins P, Schmidt S, Scott WK, et al. Neovascular age-related macular degeneration and its association with LOC387715 and complement factor H polymorphism. Arch Ophthalmol. 2007; 125(1):63-7.
8. Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125(1):55-62. Comment in: JAMA. 2007;297(4):401-2.
9. Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9): 1055-9.
10. Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102(20):7227-32. Comment in: Proc Natl Acad Sci U S A. 2005;102(20):7053-4.
11. Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006; 38(10):1173-7. Erratum in: Nat Genet. 2007;39(4):567.
12. Postel EA, Agarwal A, Caldwell J, Gallins P, Toth C, Schmidt S, et al. Complement factor H increases risk for atrophic age-related macular degeneration. Ophthalmology. 2006;113(9):1504-7.
13. Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, Ott J, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005;77(1):149-53.
14. Scott WK, Schmidt S, Hauser MA, Gallins P, Schnetz-Boutaud N, Spencer KL, et al. Independent effects of complement factor H Y402H polymorphism and cigarette smoking on risk of age-related macular degeneration. Ophthalmology. 2007;114(6):1151-6.
15. Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389-407.
16. Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227-36.
17. Schmidt S, Hauser MA, Scott WK, Postel EA, Agarwal A, Gallins P, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78(5):852-64.
18. Spencer KL, Hauser MA, Olson LM, Schmidt S, Scott WK, Gallins P, et al. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum Mol Genet. 2007;16(16):1986-92.
19. Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT; AMD Genetics Clinical Study Group, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458-62.
20. Churchill AJ, Carter JG, Lovell HC, Ramsden C, Turner SJ, Yeung A, et al. VEGF polymorphisms are associated with neovascular age-related macular degeneration. Hum Mol Genet. 2006;15(19):2955-61.
21. Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, et al. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006; 47(1):329-35.
22. Eyetech Study Group. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology. 2003;110(5):979-86. Comment on: Ophthalmology. 2003;110(5):879-81.
23. Reddy VM, Zamora RL, Kaplan HJ. Distribution of growth factors in subfoveal neovascular membranes in age-related macular degeneration and presumed ocular histoplasmosis syndrome. Am J Ophthalmol.1995;120(3):291-301.
24. Souied EH, Benlian P, Amouyel P, Feingold J, Lagarde JP, Munnich A, et al. The epsilon4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. Am J Ophthalmol.1998; 125(3):353-9.
25. Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16 Spec No. 2:R174-82.
26. den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109(1):121-4.
27. Wiggs JL, Nemesure B. Glossary of genetic terms. Arch Ophthalmol. 2007; 125:E1-E7.
28. Lorent K, Overbergh L, Moechars D, De Strooper B, Van Leuven F, Van den Berghe H. Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the alpha-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, alpha-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience. 1995;65(4):1009-25.
29. Conley YP, Thalamuthu A, Jakobsdottir J, Weeks DE, Mah T, Ferrell RE, Gorin MB. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005;14(14):1991-2002.
30. Baird PN, Guida E, Chu DT, Vu HT, Guymer RH. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45(5):1311-5.
31. Klaver CC, Kliffen M, van Duijn CM, Hofman A, Cruts M, Grobbee DE, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63(1):200-6. Erratum in: Am J Hum Genet. 1998;63(4):1252.
32. Combadière C, Feumi C, Raoul W, Keller N, Rodéro M, Pézard A, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007; 117(10):2920-8. Comment in: J Clin Invest. 2007;117(10):2758-62.
33. Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9(7):917-24. Comment in: Nat Neurosci. 2006;9(7):859-61.
34. Tuo J, Smith BC, Bojanowski CM, Meleth AD, Gery I, Csaky KG, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18(11):1297-9.
35. Ambati J, Anand A, Fernandez S, Sakurai E, Lynn BC, Kuziel WA, et al. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat Med. 2003;9(11):1390-7. Comment in: Nat Med. 2003; 9(11):1350-1. Nat Med. 2004;10(7):663; author reply 663.
36. Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204(5): 1057-69.
37. Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1999;277(5333):1805-7. Comment in: Science. 1997;277(5333): 1765-6. Science. 1998;279(5354):1107.
38. Stone EM, Webster AR, Vandenburgh K, Streb LM, Hockey RR, Lotery AJ, Sheffield VC. Allelic variation in ABCR associated with Stargardt disease but not age-related macular degeneration. Nat Genet. 1998;20(4):328-9.
39. Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, Schain M, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12(24):3315-23.
40. Farah ME, Oshima A, Costa RA, Sallum JF. Degeneração macular relacionada à idade: modalidades terapêuticas. Arq Bras Oftalmol. 2001;64(6):583-8.
 Address for correspondence:
Address for correspondence:
Luciana Almeida
Av. Conselheiro Furtado, 2.818
Belém (PA) CEP 66063-060
E-mail: [email protected]
Recebido para publicação em 18.02.2008
Última versão recebida em 02.02.2009
Aprovação em 03.03.2009
Trabalho realizado no Hospital São Geraldo e Instituto Nacional de Ciência e Tecnologia de Medicina Molecular, Laboratório de Genética Molecular, Faculdade de Medicina - UFMG - Belo Horizonte (MG) - Brazil.