
Adália Dias Dourado Oliveira1; Pedro Alves d'Azevedo2; Luciene Barbosa de Sousa4; Cristina Viana-Niero5; Waldemar Francisco6; Cláudio Lottenberg7; Marines Dalla Valle Martino7; Ana Luisa Höfling-Lima7
DOI: 10.1590/S0004-27492007000400018
ABSTRACT
PURPOSE: To evaluate different methods of oxacillin susceptibility testing of ocular isolates, considering polymerase chain reaction (PCR) as the 'gold standard', and to compare the in vitro susceptibility to oxacillin with that of other antimicrobials used in ophthalmologic practice. METHODS: The Vitek gram-positive identification card was used to identify ocular coagulase negative Staphylococcus species. The presence of the mecA gene was determined by the polymerase chain reaction assay with a combination of two primer sets (mecA and 16S rRNA) in a single region. Results were analyzed and compared with other oxacillin susceptibility methods: PBP2a detection by rapid slide latex agglutination test (SLA); oxacillin E-test; the Vitek automated gram-positive susceptibility card (GPS-105); the oxacillin salt agar screening test (OSAS) at a concentration of 6.0, 1.0 and 0.75 µg oxacillin per ml and the cefoxitin disk diffusion test (CDD). Automated susceptibility was also determined to other antimicrobial agents (fluoroquinolones, penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin and rifampin. RESULTS: Of the 69 CoNS isolates tested, 71% were mecA-positive and 29% mecA-negative. All methods tested had a statistically significant agreement with polymerase chain reaction. There was a tendency of positive polymerase chain reaction predomination among the S. epidermidis isolates in comparison to non-epidermidis isolates, although this was not statistically significant (78.3% vs. 56.5%; chi2= 2.54; P= 0.11). The oxacillin salt agar screening test (0.75 µg oxacillin/ml) showed the best performance, with 100% sensitivity and negative predictive value; 95% specificity and 98% positive predictive value. Using the E-test, the mecA-positive isolates were statistically significantly more resistant to ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin (P= 0.002; P= 0.008; P= 0.002 and P= 0.003, respectively). There was a statistically significant higher proportion of resistance of the coagulase negative Staphylococcus mecA-positives for: penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin and tetracycline (P<0.05). All coagulase negative Staphylococcus species were susceptible to vancomycin and there was no statistically significant correlation between the mecA-positive isolates and resistance to trimethoprim-sulfamethoxazole or to rifampin. CONCLUSIONS: In the present study, we found that the E-test and the oxacillin salt agar screening test S (0.75 µg oxacillin per ml), when compared with polymerase chain reaction, were the most accurate currently available methods to phenotypically detect oxacillin resistance of coagulase negative Staphylococcus species. This study demonstrated that a good option for screening of ocular isolates for oxacillin resistance in the microbiology laboratory is the cefoxitin disk diffusion test and the automated Vitek system. We believe it is important to have available methods that accurately detect methicillin resistance of the less commonly encountered species, chiefly because of their increasing importance as opportunistic pathogens.
Keywords: Methicillin resistance; Sensitivity and specificity; Latex fixation tests; Oxacillin; Microbial sensitivity tests; Staphylococcus; Staphylococcus; Coagulase; Eye infections; bacterial; Comparative study
RESUMO
OBJETIVOS: Avaliar os diferentes métodos de suscetibilidade à oxacillina, em isolados oculares, considerando a reação em cadeia da polimerase (PCR) como "padrão-ouro" e comparar a suscetibilidade in vitro para outros antimicrobianos de uso oftalmológico. MÉTODOS: O sistema automatizado Vitek foi utilizado para identificar as diferentes espécies de Staphylococcus coagulase negativo (SCoN). A presença do gene mecA foi determinado pela reação em cadeia da polimerase com a combinação de 2 "primer" sets (mecA e 16S rRNA) em uma única região. Estes resultados foram analisados e comparados com outros métodos de suscetibilidade à oxacilina: detecção da proteína PBP2a pelo teste de aglutinação em látex (SLA); E-test oxacilina; o sistema automatizado Vitek (GPS-105); o teste de triagem em ágar (OSAS) com oxacilina nas concentrações de 6,0, 1,0 e 0,75 µg oxacilina por ml e o teste de disco difusão com cefoxitina (CDD). A suscetibilidade automatizada foi obtida para os seguintes agentes antimicrobianos: fluorquinolonas, penicilina G, amoxicilina-ampicilina, cefazolina, ampicilina-sulbactam, eritromicina, clindamicina, gentamicina, tetraciclina, sulfametoxazol-trimetoprima, vancomicina e rifampicina. RESULTADOS: Dos 69 Staphylococcus coagulase negativo testados, 71% foram mecA-positivos e 29%, mecA-negativos. Todos os métodos testados apresentaram concordância estatisticamente significante com a reação em cadeia da polimerase. Houve tendência à predominância da positividade da reação em cadeia da polimerase entre os S. epidermidis comparado aos não-epidermidis, embora sem significância estatistica (78,3% vs. 56,5%; chi2= 2,54; p=0,11). O teste de triagem em ágar (0,75 µg oxacilina/ml) apresentou a melhor performance com resultados de: 100% de sensibilidade e valor preditivo negativo, 95% de especificidade e 98% de valor preditivo positivo. Os isolados mecA-positivos foram estatisticamente significativavos mais resistentes para ciprofloxacina, ofloxacina, gatifloxacina e moxifloxacina, no E-test (p=0,002; p=0,008; p=0,002 e p=0,003). Houve maior proporção estatisticamente significativa de resistência entre os Staphylococcus coagulase negativo mecA-positivos para: penicilina G, amoxicilina-ampicilina, cefazolina, ampicilina-sulbactam, eritromicina, clindamicina, gentamicina e tetraciclina. (p<=0,05) Todos os Staphylococcus coagulase negativos foram suscetíveis à vancomicina e não houve correlação estatisticamente significativa entre as amostras mecA-positivas e a resistência para sulfametoxazol-trimetoprima e rifampicina. CONCLUSÕES: No presente estudo, foi observado que o E-test e o OSAS (0,75 µg oxacilina por ml), comparado à reação em cadeia da polimerase, foram os métodos fenotípicos mais acurados em detectar a resistência à oxacilina nos Staphylococcus coagulase negativos. Foi demonstrado que os testes de disco difusão com cefoxitina e o método automatizado (Vitek) são boas opções para a triagem da resistência à oxacilina em laboratórios de microbiologia ocular. Destacou-se a importância de métodos acurados para detectar a resistência à meticilina dentre as espécies menos freqüentemente encontradas, considerando a crescente importância destes patógenos oportunistas.
Descritores: Resistência a meticilina; Sensibilidade e especificidade; Testes de fixação do látex; Oxacilina; Testes de sensibilidade microbiana; Staphylococcus; Staphylococcus; Coagulase; Infecções oculares bacterianas; Estudo comparativo
ORIGINAL ARTICLE
Laboratory detection methods for methicillin resistance in coagulase negative Staphylococcus isolated from ophthalmic infections
Métodos laboratoriais para a detecção da resistência à meticilina nos Staphylococcus coagulase negativos de infecções oculares
Adália Dias Dourado OliveiraI; Pedro Alves d'AzevedoII; Luciene Barbosa de SousaIII; Cristina Viana-NieroIV; Waldemar FranciscoV; Cláudio LottenbergVI; Marines Dalla Valle MartinoVI; Ana Luisa Höfling-LimaVI
IDepartamento de Oftalmologia, Universidade Federal de São Paulo - UNIFESP - São Paulo (SP) - Brasil
IIDepartamento de Microbiologia e Parasitologia, Fundação Faculdade Federal de Ciências Médicas de Porto Alegre - FFFCM - Porto Alegre (RS) - Brasil e Laboratório Especial de Microbiologia Clínica, Divisão de Doenças Infecciosas da UNIFESP - São Paulo (SP) - Brasil
IIIDepartamento de Oftalmologia da UNIFESP - São Paulo (SP) - Brasil
IVDepartamento de Microbiologia, Imunologia da UNIFESP - São Paulo (SP) - Brasil
VDepartamento de Patologia, Faculdade de Ciências Médicas da Santa Casa de Misericórdia de São Paulo (SP) - Brasil
VIHospital Israelita Albert Einstein - HIAE - São Paulo (SP) - Brasil
ABSTRACT
PURPOSE: To evaluate different methods of oxacillin susceptibility testing of ocular isolates, considering polymerase chain reaction (PCR) as the 'gold standard', and to compare the in vitro susceptibility to oxacillin with that of other antimicrobials used in ophthalmologic practice.
METHODS: The Vitek gram-positive identification card was used to identify ocular coagulase negative Staphylococcus species. The presence of the mecA gene was determined by the polymerase chain reaction assay with a combination of two primer sets (mecA and 16S rRNA) in a single region. Results were analyzed and compared with other oxacillin susceptibility methods: PBP2a detection by rapid slide latex agglutination test (SLA); oxacillin E-test; the Vitek automated gram-positive susceptibility card (GPS-105); the oxacillin salt agar screening test (OSAS) at a concentration of 6.0, 1.0 and 0.75 µg oxacillin per ml and the cefoxitin disk diffusion test (CDD). Automated susceptibility was also determined to other antimicrobial agents (fluoroquinolones, penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin and rifampin.
RESULTS: Of the 69 CoNS isolates tested, 71% were mecA-positive and 29% mecA-negative. All methods tested had a statistically significant agreement with polymerase chain reaction. There was a tendency of positive polymerase chain reaction predomination among the S. epidermidis isolates in comparison to non-epidermidis isolates, although this was not statistically significant (78.3% vs. 56.5%; c2= 2.54; P= 0.11). The oxacillin salt agar screening test (0.75 µg oxacillin/ml) showed the best performance, with 100% sensitivity and negative predictive value; 95% specificity and 98% positive predictive value. Using the E-test, the mecA-positive isolates were statistically significantly more resistant to ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin (P= 0.002; P= 0.008; P= 0.002 and P= 0.003, respectively). There was a statistically significant higher proportion of resistance of the coagulase negative Staphylococcus mecA-positives for: penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin and tetracycline (P<0.05). All coagulase negative Staphylococcus species were susceptible to vancomycin and there was no statistically significant correlation between the mecA-positive isolates and resistance to trimethoprim-sulfamethoxazole or to rifampin.
CONCLUSIONS: In the present study, we found that the E-test and the oxacillin salt agar screening test S (0.75 µg oxacillin per ml), when compared with polymerase chain reaction, were the most accurate currently available methods to phenotypically detect oxacillin resistance of coagulase negative Staphylococcus species. This study demonstrated that a good option for screening of ocular isolates for oxacillin resistance in the microbiology laboratory is the cefoxitin disk diffusion test and the automated Vitek system. We believe it is important to have available methods that accurately detect methicillin resistance of the less commonly encountered species, chiefly because of their increasing importance as opportunistic pathogens.
Keywords: Methicillin resistance; Sensitivity and specificity; Latex fixation tests; Oxacillin; Microbial sensitivity tests; Staphylococcus/isolation & purification; Coagulase; Eye infections, bacterial; Comparative study
RESUMO
OBJETIVOS: Avaliar os diferentes métodos de suscetibilidade à oxacillina, em isolados oculares, considerando a reação em cadeia da polimerase (PCR) como "padrão-ouro" e comparar a suscetibilidade in vitro para outros antimicrobianos de uso oftalmológico.
MÉTODOS: O sistema automatizado Vitek foi utilizado para identificar as diferentes espécies de Staphylococcus coagulase negativo (SCoN). A presença do gene mecA foi determinado pela reação em cadeia da polimerase com a combinação de 2 "primer" sets (mecA e 16S rRNA) em uma única região. Estes resultados foram analisados e comparados com outros métodos de suscetibilidade à oxacilina: detecção da proteína PBP2a pelo teste de aglutinação em látex (SLA); E-test oxacilina; o sistema automatizado Vitek (GPS-105); o teste de triagem em ágar (OSAS) com oxacilina nas concentrações de 6,0, 1,0 e 0,75 µg oxacilina por ml e o teste de disco difusão com cefoxitina (CDD). A suscetibilidade automatizada foi obtida para os seguintes agentes antimicrobianos: fluorquinolonas, penicilina G, amoxicilina-ampicilina, cefazolina, ampicilina-sulbactam, eritromicina, clindamicina, gentamicina, tetraciclina, sulfametoxazol-trimetoprima, vancomicina e rifampicina.
RESULTADOS: Dos 69 Staphylococcus coagulase negativo testados, 71% foram mecA-positivos e 29%, mecA-negativos. Todos os métodos testados apresentaram concordância estatisticamente significante com a reação em cadeia da polimerase. Houve tendência à predominância da positividade da reação em cadeia da polimerase entre os S. epidermidis comparado aos não-epidermidis, embora sem significância estatistica (78,3% vs. 56,5%; c2= 2,54; p=0,11). O teste de triagem em ágar (0,75 µg oxacilina/ml) apresentou a melhor performance com resultados de: 100% de sensibilidade e valor preditivo negativo, 95% de especificidade e 98% de valor preditivo positivo. Os isolados mecA-positivos foram estatisticamente significativavos mais resistentes para ciprofloxacina, ofloxacina, gatifloxacina e moxifloxacina, no E-test (p=0,002; p=0,008; p=0,002 e p=0,003). Houve maior proporção estatisticamente significativa de resistência entre os Staphylococcus coagulase negativo mecA-positivos para: penicilina G, amoxicilina-ampicilina, cefazolina, ampicilina-sulbactam, eritromicina, clindamicina, gentamicina e tetraciclina. (p<=0,05) Todos os Staphylococcus coagulase negativos foram suscetíveis à vancomicina e não houve correlação estatisticamente significativa entre as amostras mecA-positivas e a resistência para sulfametoxazol-trimetoprima e rifampicina.
CONCLUSÕES: No presente estudo, foi observado que o E-test e o OSAS (0,75 µg oxacilina por ml), comparado à reação em cadeia da polimerase, foram os métodos fenotípicos mais acurados em detectar a resistência à oxacilina nos Staphylococcus coagulase negativos. Foi demonstrado que os testes de disco difusão com cefoxitina e o método automatizado (Vitek) são boas opções para a triagem da resistência à oxacilina em laboratórios de microbiologia ocular. Destacou-se a importância de métodos acurados para detectar a resistência à meticilina dentre as espécies menos freqüentemente encontradas, considerando a crescente importância destes patógenos oportunistas.
Descritores: Resistência a meticilina; Sensibilidade e especificidade; Testes de fixação do látex; Oxacilina; Testes de sensibilidade microbiana; Staphylococcus/isolamento & purificação; Coagulase; Infecções oculares bacterianas; Estudo comparativo
INTRODUCTION
Coagulase-negative Staphylococcus (CoNS) species are commonly isolated bacteria, often admixed with more typical ocular microbes, and can lead to major infections, including keratitis, conjunctivitis and endophthalmitis(1-3). Staphylococcus epidermidis, the predominant species of the CoNS, is the most commonly cultured intraocular pathogen, and accounts for an average of 40% postoperative and posttraumatic endophthalmitis cases(4-5).
Methicillin-resistant coagulase negative Staphylococcus (MRCoNS) is just one species of intraocular pathogens(6) and one report in the literature has described patients with MRCoNS among the preoperative ones seen at a Japanese eye clinic(7). Resistance to methicillin is associated with in vitro crossing resistance to other antimicrobials(6,8). The laboratory data from the Ophthalmological Department of the University of São Paulo (UNIFESP) has documented a recent increase in infections due to this microorganism(9). In contrast to infection at other sites, the ocular therapeutic difficulties posed by the MRCoNS have been virtually unrecognized.
Two basic mechanisms are responsible for the resistance of staphylococci to the beta-lactamic antimicrobials: first, the b-lactamase production that destroys these agents, and, second, alteration of proteins located in the cellular wall of the bacteria, the so-called penicillin-binding proteins (PBPs)(10). Most resistance to oxacillin by staphylococci is mediated by the mecA gene, which codes for the production of a supplemental penicillin-binding protein, PBP2a or 2', which is expressed either homogeneously or heterogeneously(11-12). PBP2a has a low affinity for beta-lactamic antimicrobials. Homogeneous resistance is easily detected with standard testing methods, whereas heterogeneous expression is more difficult to detect with some methods, because only a fraction of the population (e.g., 1 in 100,000 cells) expresses the resistance phenotype(13).
Tests for mecA or for the protein encoded by mecA, PBP2a, are the most accurate methods for prediction of resistance to oxacillin, and could be used to confirm results of staphylococci isolates from serious infections(14). Oxacillin is no longer the agent recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee of Clinical and Laboratory Standards - NCCLS) for CoNS phenotypic tests to predict resistance to penicillinase-stable penicillins (PSPs). Antimicrobial susceptibility tests using oxacillin are often difficult to interpret, despite changes that have improved discrimination between oxacillin-susceptible and -resistant strains. Minimum inhibitory concentration (MIC) panels in which oxacillin is tested must be examined carefully to detect any growth that may be indicative of resistance(13-16). Several groups of investigators(17-19) have reported that the results of cefoxitin (30 µg) disk diffusion tests correlate better with the presence of mecA than do the results of disk diffusion tests using oxacillin, and the cefoxitin disk test is now the preferred method for testing CoNS(13).
The oxacillin salt agar screen (OSAS) has been recommended by CLSI (2005) as an additional test that can be used if the dilution tests or disk diffusion tests are indeterminate. However, the agar screen test is recommended only for S. aureus, and it also can be difficult to read, especially if the isolate is heteroresistant(20-21).
The purpose of the study described herein was to evaluate different oxacillin susceptibility methods currently used for ocular isolates, considering the multiplex PCR assay for detection of the mecA gene to be the 'gold standard', and to compare the susceptibility of mecA-positive CoNS with mecA-negative CoNS to other antimicrobials used in ophthalmic practice: fluoroquinolones, penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin and rifampin.
METHODS
Bacterial isolates and bacteriologic methods
Fresh and cryopreserved bacterial isolates were used. They were collected between July 2002 and July 2004 and processed according to standard microbiologic protocols in a reference ophthalmological laboratory. Samples included 69 CoNS isolates from clinically diagnosed keratitis, conjunctivitis or endophthalmitis. The laboratory had identified the organisms as CoNS by colony morphology, Gram stain characteristics, and by catalase and coagulase tests. S. aureus (ATCC 25923) was used as the mecA gene negative control and S. aureus (ATCC 43300) was used as the mecA gene positive control.
Inoculum preparation
Inocula for all tests were prepared using trypicase soy agar (TSA) (Difco, Le Pont de Claix, France) supplemented with 5% sheep blood that had been streaked with a single colony from an initial subculture plate and incubated for 18 to 24 h. The test inoculum was prepared by removing growth from the agar plate, inoculating it directly into Mueller-Hinton broth (MHB) (Difco, Le Pont de Claix, France), and adjusting the inoculum to equal a 0.5 McFarland turbidity standard.
Identification of species
The Vitek (BioMerieux, Hazelwood, MI) gram-positive identification card was used to identify the CoNS species. Card inoculation, sealing, incubation, and reading were performed according to the instructions of the manufacturer. The species were then classified into two groups: epidermidis (S. epidermidis CoNS) and non-epidermidis (CoNS non-S. epidermidis).
Polymerase chain reaction (PCR) detection of mecA gene
The mecA gene, in combination with the 16S rRNA gene, was detected with a PCR technique based on the procedure described previously(22). The multiplex assay combined two primer sets (mecA and 16S rRNA) in a single region, and presence of the mecA gene was considered the 'gold standard' for oxacillin resistance.
PBP2a latex slide agglutination test (LSA)
Penicillin-binding protein 2a detection by the rapid slide latex agglutination test (Slidex MRSA Detection Test, BioMerieux, Paris, France) was performed according to the manufacturer's instructions with slight modification: the resulting agglutination pattern was read at 10. Bacterial cells (approximately 5 µl) were obtained from a fresh subculture and suspended in 4 drops of extraction reagent 1 and boiled for 3 min. The suspension was allowed to cool to room temperature, after which 1 drop of extraction reagent 2 was added, and the mixture vortexed thoroughly. The suspension was then centrifuged at 1500 x g for 5 min. A 50 µl aliquot of the supernatant was mixed with 1 drop of anti-PBP2a monoclonal antibody-sensitized latex beads. A negative control test was performed by using 50 µl of supernatant mixed with 1 drop of negative control latex. The isolates were then placed on a shaker and gently mixed for up to 15 mins.
Automated susceptibility test (AS)
The AS test was performed with the Gram-Positive Susceptibility card (GPS-105) of the Vitek system (BioMerieux, Hazelwood, MI), according to the manufacturer's recommendation. The susceptibility of all CoNS isolates was determinated for oxacillin and the other antimicrobials used in ophthalmology: penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin and rifampin.
Oxacillin salt agar screen susceptibility test (OSAS)
Agar screen tests for susceptibility to oxacillin were performed by a method described previously(23). The OSAS test was performed with agar plates produced by PROBAC (São Paulo, Brazil). A total of 104 CFU was spot inoculated onto Mueller-Hinton agar (MHA) (PROBAC, São Paulo, Brazil) with 4% NaCl supplementation containing 6, 1 or 0.75 µg of oxacillin per ml. Plates were read after a 24 h incubation at 35ºC. If any colonies of growth were detected, the test was considered to be positive for resistance.
Disk susceptibility test (DD)
The CLSI reference method for disk diffusion was used to test cefoxitin (30 µg) (Oxoid, Basingstore, England)(13). MHA plates were incubated at 35ºC, and zone diameters were read at 18 h to 24 h.
E-test
The minimum inhibitory concentrations (MIC) for oxacillin, ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin were each determinated using the E-test (AB Biodisk, Solna, Sweden). Plates were incubated at 35ºC for 18 to 24 hours. The MIC value (E-test) was read at the point where the edge of the growing culture intersects the strip. The antimicrobial susceptibility of each bacterial isolate was determined by comparing the MIC with that of the Clinical and Laboratory Standards Institute (CLSI)(14) when available. Each isolate was classified as susceptible or nonsusceptible (intermediate or resistant).
b-Lactamase test
b-Lactamase production was identified using nitrocefin disks (Difco, Le Pont de Claix, France). This test was performed with all mecA-negative isolates that were resistant by E-test or by the automated method.
Amoxicillin-clavulanic acid disk diffusion test
The amoxicillin-clavulanic acid disk test (20 µg amoxicillin and 10 µg clavulanic acid) (Oxoid,Basingstore, England) was performed with all mecA-negative isolates that were found to be resistant by E-test or by the automated method. The breakpoint for susceptibility was a zone of inhibition >20 mm in diameter after 24 h of incubation at 30ºC.
Data analysis
Evaluation of data comparing results of the mecA gene PCR assay and results from the other susceptibility tests required a matched 2-by-2 table. Evaluation of data was calculated using the SPSS (Statistical Package for Social Sciences, 11.0 version, SPSS Inc, Chigaco). Difference in susceptibility methods and significance of the results was calculated by the Chi-square test or Fisher exact test. Statistical significance was accepted when the P-value was <0.05. Validity tests (sensitivity, specificity, positive predictive value and negative predictive value) were also calculated. Sensitivity was defined as the percentage of mecA-positive isolates determined to be nonsusceptible by phenotypic testing, and specificity was defined as the percentage of mecA-negative isolates determined to be susceptible by phenotypic testing. The isolates were designated as susceptible or nonsusceptible (resistant and intermediate) based on the 2006 CLSI document.
RESULTS
Of 69 CoNS tested, 49 (71%) were mecA-positive and 20 (29%) were mecA-negative. They were classified into one of two groups based on species identification: 46 (66.7%) were S. epidermidis and 23 (33.3%) were non-epidermidis (9 S. auricularis; 4 S. haemolyticus; 4 S. capitis; 2 S. simulans; 1 S. hominis; 1 S. sciuris, 1 S. cohnii and 1 S. saprophyticus). For the mecA-negative isolates only the 16S rRNA specific band was observed.
All methods tested had a statistically significant agreement with PCR (Table 1). Table 2 shows the results of oxacillin susceptibility tests for each of the 69 CoNS isolates.
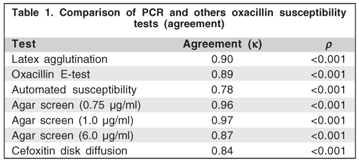
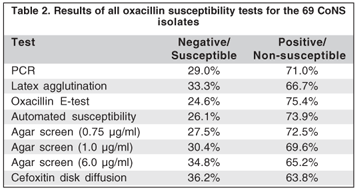
All mecA-positive isolates were oxacillin resistant by the E-test, although three isolates that were resistant by the E-test did not show the mecA gene. In the automated method, four resistant isolates did not show the mecA gene, although they were positive by the nitrocefn disk test and were susceptible to amoxicillin-clavulonic acid disks. Only 46 mecA-positives showed latex agglutination (k=0.90; P<0.001). All mecA-positives were detected by the agar screen test with 0.75 oxacillin mg/ml, although, one and six isolates had not being detected by the agar screen test with 1 and 6 oxacillin mg/ml respectively (k=0.96; k=0.97;k=0.87; P<0.001 respectively). The cefoxitin disk diffusion test did not show resistance in 5 mecA-positive isolates (k=0.84; P<0.001).
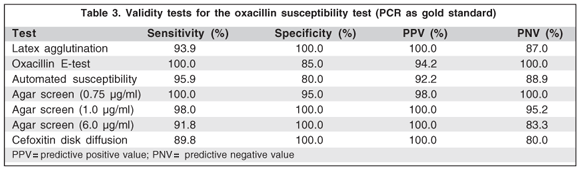
Validity of results for all methods tested is shown in table 3. The agar screen oxacillin test (0.75 µg/ml) showed the best performance with results of 100% sensitivity and negative predictive value, and 95% specificity and 98% positive predictive value. When comparing the epidermidis with the non-epidermidis groups, the latter had the worst values regarding sensitivities in the latex, automated, agar screen (6 and 1 µg/ml) and cefoxitin disk diffusion tests, and in specificities of the E-test and agar screen (0.75 µg/ml) tests (Table 4).
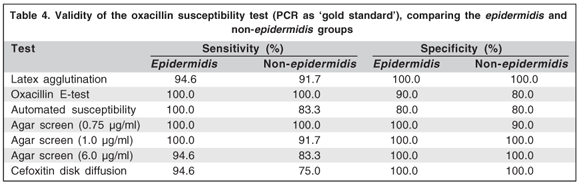
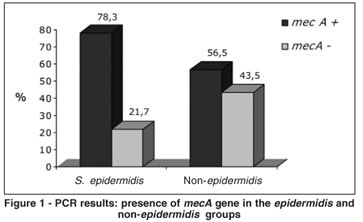
There was a tendency toward mecA predominance among the S. epidermidis compared to non-epidermidis groups, although the difference was not statistically significant (78.3% vs. 56.5%; µ2=2.54; P = 0.11) (Figure 1). For the non-epidermidis group, there were more false-positives in the E-test, automated method and the agar screen test with 0.75 oxacillin mg/ml, and more false-negatives in the automated method, cefoxitin disk diffusion test and the the agar screen test with 1 oxacillin µg/ml than for the epidermidis group. For the epidermidis group, there were more false negatives for the latex agglutination test (Table 5).
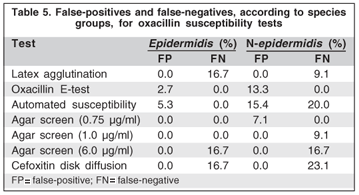
By the E-test, the mecA-positive isolates were statistically significantly more resistant to ciprofloxacin, ofloxacin, gatifloxacin and moxifloxacin (P=0.002; P=0.008; P=0.002 and P=0.003) (Figure 2).
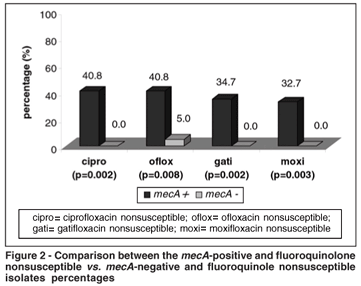
There was a statistically significant higher proportion of resistance of the CoNS mecA-positives to penicillin G, amoxicillin-ampicillin, cefazolin, ampicillin-sulbactam, erythromycin, clindamycin, gentamicin and tetracycline (P<0.05). Resistance to vancomycin was not observed. There was no statistically significant correlation between the mecA-positive isolates and resistance for trimethoprim-sulfamethoxazole or rifampin (Figura 3).
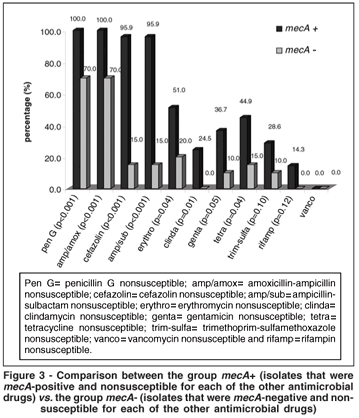
In comparing the two species groups, there was a statistically significant higher proportion of resistance of the S. epidermidis than of the non-epidermidis groups to cefazolin and to ampicillin-sulbactam (P=0.05 and P=0.05, respectively); the S. epidermidis isolates were also more resistant to fluoroquinolones, penicillin G, amoxicillin-ampicillin, erythromycin, clindamycin, gentamicin, trimethoprim-sulfamethoxazole and rifampin (although with no statistical significance).
DISCUSSION
Staphylococci are the most commonly pathogens isolated from ocular tissues(1-2,5,24). Because of their ubiquitous nature and relatively low virulence, staphylococci, other than S. aureus, in the past were often simply reported by the microbiology laboratory as CoNS. However, over the past 15 years, there has been increased documentation of ocular infections caused by CoNS(1,5-6,9,25-26). Because the antimicrobial susceptibility of CoNS is unpredictable, and because multiresistant isolates are common, the recommendation now is to perform antimicrobial susceptibility testing in all cases of clinically significant ocular infections caused by these organisms.
Identification of methicillin-resistant staphylococci in the laboratory is complicated by the heterogeneous nature of the resistance and by the variables that influence its expression (i.e., medium, inoculum size, pH, temperature, and salt concentration)(27-28). Only a few cells in a population of bacteria may be PBP2a positive because of heterogeneity of mecA gene expression, especially in CoNS(29). Furthermore, methicillin resistance can involve non-PBP2a-dependent mechanisms, such as hyperproduction of b-lactamase(30).
Several studies have demonstrated that PCR is a sensitive method for the detection of methicillin resistance in CoNS(22,31). Unfortunately, most laboratories, especially ophthalmologic ones, are not in a position to perform this test. The percentage of mecA-positive cultures and the predominance of S. epidermidis in the eye was similar to findings at other sites(29,32-37). A multicenter study in Brazil showed methicillin resistance in 87.7% of CoNS isolated from infections of the bloodstream(38).
Previous studies that examined results of the GPI card (automated Vitek system), have reported CoNS identification to vary from 67.3% to 89%. These discrepant results in identification are related to species that are less commonly isolated, while the system was quite reliable for the most commonly isolated and more clinically relevant species: S. epidermidis(37,39-42).
All of the oxacillin susceptibilities tests performed in this study were in overall agreement with PCR results, and the phenotypic methods evaluated in the present study also appeared to perform well for the detection of oxacillin resistance.
The LSA test was done with extension of the reaction time to 10 min, with resultant 93.9% sensitivity; this is not as good a result as that reported in the literature, which describes the use of oxacillin-induced colonies(34,43) or the use of a greater concentration of inoculum(44-45). This test has the advantage of having a short turnaround time, but it was the most expensive test performed and failed to detect oxacillin resistance in 3 (6.1%) mecA-positive CoNS isolates.
The cefoxitin disk test is rapidly becoming the preferred method for for the detection of oxacillin heteroresistance(19,46-47). This test is easier to read, has higher specificity than and equal sensitivity to the oxacillin disk test for CoNS(46). The specificity in this study was 100%; the sensitivity, however, although similar to that reported in the literature (range, 85% to 96%)(19), showed a decrease in overall percentage rate because results for the non-epidermidis group were worse than were those for the epidermidis group.
In this version of the automated test, detection of methicillin resistance was based solely on determination of oxacillin MIC. The literature reports high sensitivities (range, 95.7% to 100%) but only a moderate degree of specificity (range, 61% to 91.8%) for the Vitek system (first version) with CoNS, similar to our results(34,44-45,48-49) . Compared to the more conventional phenotypic methods, which can take up to 24 h, the automated test provides results in about 10 hs, which may have a potential impact on the optimal management of CoNS infections. Additionally, it is easy to use and is relatively inexpensive because of reduced laboratory expenses and other general costs. The reduced specificity encountered in our study may be attributable to the high oxacillin MICs for mecA-negative non-epidermidis isolates(49). The MIC breakpoints were found to be less accurate when they were applied to some species of CoNS, and our findings confirm this observation(49) (Table 3). Four of the false-positive isolates were b-lactam hyperproducers, which was confirmed by the nitrocefin and amoxicillin-clavulonic acid disk diffusion tests.
Although it has been reported(27) that OSAS (6.0 µg/ml) is ineffective for CoNS, the OSAS (0.75 µg/ml) in the present study showed the best performance of all of the used tests, including the other two concentrations (1.0 and 6.0 µg/ml), and it could thus be considered an accurate method for confirmation of resistance. Comparative studies to assess the OSAS and disk diffusion test have shown that OSAS has good sensitivity for the identification of methicillin-resistant strains(29,33,36,45,50-52).
The E-test and OSAS (0.75 µg/ml) were equally reliable in their detection of mecA-positive isolates. However, these methods proved to be less accurate in discerning isolates that lacked the mecA gene. Louie et al.(45) found a relatively low level of specificity (78%) for the E-test, similar to what we noted. By the E-test and OSAS oxacillin (0.75 µg/ml), 3 (1 S. epidermidis, 1 S. auricularis and 1 S. haemolyticus) of 20 (15%) and 1 (S. auricularis) of 20 (5%) mecA-negative isolates appeared to be resistant. Again, the new breakpoints values, reported by Tenover et al.(27), in 1999, were chosen because they were the best for maximizing the sensitivity of detection of mecA-positive S. epidermidis isolates without severely compromising specificity. However, for CoNS other than S. epidermidis, the actual breakpoints are less effective in differentiating mecA-positive from mecA-negative isolates. Rowe et al.(23), indicated that, for S. epidermidis, the E-test and the OSAS (0.6 µg/ml) correctly discriminated mecA-positive from mecA-negative isolates with 100% sensitivity. However, these tests were not satisfactory to assess methicillin resistance to other CoNS species. Our study showed 100% sensitivity of the E-test and OSAS (0.75 µg/ml) for both groups (epidermidis and non-epidermidis), but there were differences in OSAS 1 and 6 µg/ml for the two groups (100% vs. 91.7% and 94.6% vs. 83.3%, respectively). There were also differences in the latex agglutination, automated and cefoxitin disk diffusion tests. The present study used only 23 of the non-epidermidis CoNS species, and a greater number would be required for validation of the interpretation.
The false-negative results in the tests evaluated may be due to an extremely heterogeneous expression of resistance(53). On the other hand, the false-positive results may have been the result of overproduction of penicillinase, or of superexpression/alterations of constitutive PBPs. It is known that penicillinase-resistant penicillins may show some degree of lysis when such enzymes are present. The superexpression or alteration of constitutive PBPs generates a higher concentration of free transpeptidase, which synthesize the cell wall of the bacteria(53). The expression of resistance is enhanced by passage in b-lactam antimicrobials, because the susceptible subpopulation is eliminated and the highly resistant subpopulation is selected out. These antimicrobial-selected cells are therefore more uniformly resistant than is the parent strain, but the trait is unstable. With repeated subculturing in drug-free medium, the culture reverts to its heterogeneous pattern of resistance(52). Attempts to increase sensitivity for detection of methicillin resistance may do so at the cost of reduced specificity. Factors that enhance such resistance (48-h incubation, 5% NaCl, 30ºC incubation, and high doses of inoculum) may also enhance b-lactamase production(52).
Methicillin-resistant staphylococci are typically resistant to a variety of antimicrobials, including quinolones, tobramycin, clindamycin, trimethoprim-sulfamethoxazole, tetracycline, and erythromycin(52,54). Therefore, before any drug other than vancomycin is used, susceptibility of the isolate must be confirmed(24). Hussain et al.(34), in 2002, affirmed that CoNS with the mecA gene were resistant to erythromycin, clindamycin and co-trimoxazole, but that tetracycline was equally active against mecA-positive and mecA-negative isolates; no resistance was observed for vancomycin(34). It has also been reported that in vitro susceptibility of CoNS to fluoroquinolone antimicrobials is higher for the methicillin-resistant group then for the methicillin-susceptible group, and both groups of staphylococci are more susceptible to gatifloxacin and moxifloxacin than to ciprofloxacin and ofloxacin(55). The data show higher potency of the fourth generation of fluoroquinolones to treat these infections than that of the second generation(56-58).
CONCLUSION
In conclusion, because the E-test and the OSAS using 0.75 µg oxacillin per ml, compared with PCR, were the most accurate available phenotypic methods of detecting oxacillin resistance in CoNS ocular isolates, we recommend these tests to confirm results on ocular specimens, since many laboratories, especially those dedicated to ophthalmologic purposes cannot afford to perform PCR. A good option for the screening of oxacillin resistance is the cefoxitin disk diffusion test and the automated test. The differences observed in the oxacillin susceptible tests for CoNS may be explained by the fact that CLSI/NCCLS does not take into consideration the species of the isolated bacterium. Because of their increasing importance, clinically significant CoNS should be identified to the species level and should have their antimicrobial susceptibility profiled. Further studies also may better define the breakpoints for methicillin resistance in CoNS other than S. epidermidis. These steps should improve our knowledge of the role played in clinical ophthalmology diseases by this group of bacteria, and aid clinicians in the treatment and alleviation of patient suffering and morbidity. The generic identification "coagulase negative staphylococci" may be inappropriate, as it 'lumps' together species that respond differently to the same experimental conditions with respect to mecA gene detection. Not surprisingly, mecA-positive CoNS were more resistant to the other antimicrobials than were mecA-negative CoNS that are widely used in ophthalmology and were tested in the present study.
REFERENCES
1. Fukuda M, Ohashi H, Matsumoto C, Mishima S, Shimomura Y. Methicillin-resistant Staphyloccocus aureus and methicillin-resistant coagulase-negative Staphylococcus ocular surface infection efficacy of chloramphenicol eye drops. Cornea. 2002;21(7 Suppl):S86-9.
2. Ta CN, Chang RT, Singh K, Egbert PR, Shriver EM, Blumenkranz MS, et al. Antibiotic resistance patterns of ocular bacterial flora: a prospective study of patients undergoing anterior segment surgery. Ophthalmology. 2003;110 (10):1946-51.
3. Trindade RC, Bonfim AC, Resende MA. Conjunctival microbiota flora of clinically normal person who work in a hospital environment. Braz J Microbiol. 2000;31(1):12-6.
4. Davis JL, Koidou-Tsiligianni A, Pflugfelder SC, Miller D, Flynn HW, Forster RK. Coagulase-negative staphylococcal endophthalmitis. Increase in antimicrobial resistance. Ophthalmology. 1988;95(10):1404-10.
5. Wong TY, Chee SP. The epidemiology of acute endophthalmitis after cataract surgery in Asian population. Ophthalmology. 2004;111(4):699-705. Comment in: Ophthalmology. 2004;111(4):699-705.
6. Lambert SR, Stern WH. Methicillin- and gentamicin-resistant Staphylococcus epidermidis endophthalmitis after intraocular surgery. Am J Ophthalmol. 1985;99(6):725-6.
7. Kato T, Hayasaka S. Methicillin-resistant Staphyloccocus aureus and methicillin-resistant coagulase-negative staphylococci from conjunctivas of preoperative patients. Jpn J Ophthalmol. 1998;42(6):461-5.
8. Goodman DF, Gottsch JD. Methicillin-resistant Staphylococcus epidermidis keratitis treated with vancomycin. Arch Ophthalmol. 1988;106(11):1570-1.
9. Gayoso MF, Oliveira AD, d'Azevedo PA, Yu MC, Hofling-Lima AL, Francisco W. Suscetibilidade antimicrobiana in vitro dos Staphylococcus coagulase negativa oculars. Arq Bras Oftalm. In press.
10. Mulligan ME, Murray-Leisure KA, Ribner BS, Standiford HC, John JF, Korvick JA, et al. Methicillin-resistant Staphyloccocus aureus: a consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Am J Med. 1993;94(3):313-28. Comment in: Am J Med. 1995;98(6):599-600.
11. Hartman BJ, Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphyloccocus aureus. Antimicrob Agents chemother. 1986;29(1):85-92.
12. Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphyloccocus aureus. Antimicrob Agents Chemother. 1985;28(3):397-403.
13. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests, 9th ed. Wayne, PA: CLSI; 2006. (document M2-A9. 26, Nº. 1).
14. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 6th informational Supplement. Wayne, PA: CLSI; 2006 (document M100-16. 26, Nº. 3).
15. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 7th ed. Wayne, PA: CLSI; 2006. (document M7-A7. 26, Nº.2).
16. Barry AL, Jones RN. Reliability of high-content disks and modified broth dilution tests for detecting staphylococcal resistance to the penicilinase-resistant penicillins. J Clin Microbiol. 1987;25(10):1987-901.
17. Cauwelier B, Gordts B, Descheemaecker P, Van Landuyt H. Evaluation of a disk diffusion method with cefoxitin (30 microg) for detection of methicillin-resistant Staphyloccocus aureus. Eur J Clin Microbiol Infect Dis. 2004;23(5):389-92. Comment in: Eur J Clin Microbiol Infect Dis. 2004;23(11):867-8.
18. Boutiba-Ben Boubaker I, Ben Abbes R, Ben Abdallah H, Mamlouk K, Mahjoubi F, Kammoun A, et al. Evaluation of a cefoxitin disk diffusion test for the routine detection of methicillin-resistant Staphyloccocus aureus. Clin Microbiol Infect. 2004;10(8):762-5.
19. Swenson JM, Tenover FC. Cefoxitin Disk Study Group. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J Clin Microbiol. 2005;43(8):3818-23.
20. Cavassini M, Wegner A, Jaton K, Blanc DS, Bille J. Evaluation of MRSA-Screen, a simple anti-PBP 2a slide latex agglutination kit, for rapid detection of methicillin resistance in Staphyloccocus aureus. J Clin Microbiol. 1999;37 (5):1591-4. Comment in: J Clin Microbiol. 1999;37(11):3783-4.
21. Swenson JM, Williams PP, Killgore G, O'Hara CM, Tenover FC. Performance of eight methods, including two new rapid methods, for detection of oxacillin resistance in a challenge set of Staphylococcus aureus organisms. J Clin Microbiol. 2001;39(10):3785-8.
22. Geha DJ, Uhl JR, Gustaferro CA, Persing DH. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32(7):1768-72.
23. Rowe F, Vargas Superti S, Machado Scheibe R, Dias CG. Agar diffusion, agar dilution, E test, and agar screening test in the detection of methicillin resistance in staphylococci small star, filled. Diagn Microbiol Infect Dis. 2002;43 (1):45-8.
24. Pinna A, Zanetti S, Sotgiu M, Sechi LA, Fadda G, Carta F. Identification and antibiotic susceptibility of coagulase negative staphylococci isolated in corneal/external infections. Br J Ophthalmol. 1999;83(7):771-3. Comment in: Br J Ophthalmol. 2000;84(2):229.
25. Puliafito CA, Baker AS, Haaf J, Foster CS. Infectious endophthalmitis. Review of 36 cases. Ophthalmology. 1982;89(8):921-9.
26. Chalita MR, Hofling-Lima AL, Paranhos A, Jr., Schor P, Belfort R. Shifiting trends in vitro antibiotic susceptibilities for common ocular isolates during a period of 15 years. Am J Ophthalmol. 2004;137(1):43-51.
27. Tenover FC, Jones RN, Swenson JM, Zimmer B, McAllister S, Jorgensen JH. Methods for improved detection of oxacillin resistance in coagulase-negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37(12):4051-8.
28. Archer Gl, Climo MW. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob Agents Chemother. 1994;38(10):2231-7.
29. Hussain Z, Stoakes L, Lannigan R, Longo S, Nancekivell B. Evaluation of screening and commercial methods for detection of methicillin resistance in coagulase-negative staphylococci. J Clin Microbiol. 1998;36(1):273-4.
30. Bush K. Beta-lactamase inhibitors from laboratory to clinic. Clin Microbiol Rev. 1988;1(1):109-23.
31. Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29(10):2240-4.
32. Ferreira RB, Nunes AP, Kokis VM, Krepsky N, Fonseca LS, Bastos Mdo C, et al. Simultaneous detection of the mecA and ileS-2 genes in coagulase-negative staphylococci isolated from Brazilian hospitas by multiplex PCR. Diagn Microbiol Infect Dis. 2002;42(3):205-12.
33. Ferreira RB, Iorio NL, Malvar KL, Nunes AP, Fonseca LS, Bastos CC et al. Coagulase-negative staphylococci: comparison of phenotypic and genotypic oxacillin susceptibility tests and evaluation of the agar screening test by using different concentrations of oxacillin. J Clin Microbiol. 2003;41(8):3609-14. Erratum; J Clin Microbiol. 2004;42(8);3913.
34. Hussain Z, Soakes L. John Ma, Garrow S, Fitzgerald V. Detection of methicillin resistance in primary blood culture isolates of coagulase-negative staphylococci by PCR, slide agglutination, disk diffusion, and a commercial method. J Clin Microbiol. 2002;40(6):2251-3.
35. Hederstierna-Johnsen T, Schonheyder HC, Paulsen K. Detection of methicillin resistance in coagulase-negative staphylococci by cefoxitin disc diffusion and oxacillin E test. A study of consecutive bacteraemia isolates. APMIS. 2005;113 (10):688-92.
36. Caierão J, Musskopf M, Superti S, Roesch E, Dias CG, d'Azevedo PA. Evaluation of phenotypic methods for methicillin resistance characterization in coagulase-negative staphylococci (CNS). J Med Microbiol. 2004;53(Pt 12):1195-9.
37. Grasmick AE, Naito N, Bruckner DA. Clinical comparison of the AutoMicrobic system gram-positive identification card, API Staph-Ident, and conventional methods in the identification of coagulase-negative Staphylococcus spp. J Clin Microbiol. 1983;18(6):1323-8.
38. Sader HS, Sampaio JL, Zocoli C, Jones RN. Results of the 1997 SENTRY Antimicrobial Surveillance Program in Three Brazilian Medical Centers. Braz J Infect Dis. 1999;3(2):63-79.
39. Ruoff KL, Ferraro MJ, Jerz ME, Kissling J. Automated identification of gram-positive bacteria. J Clin Microbiol. 1982;16(6):1091-5.
40. Almeida RJ, Jorgensen JH, Johnson JE. Evaluation of the automicrobic system gram-positive identification card for species identification of coagulase-negative staphylococci. J Clin Microbiol. 1983;18(2):438-9.
41. Bannerman TL, Kleeman KT,Kloos WE. Evaluation of the Vitek systems gram-positive identification card for species identification of coagulase-negative staphylococci. J Clin Microbiol. 1993;31(5):1322-5.
42. Caierão J, Superti S, Dias CA, d'Azevedo PA. Automated systems in the identification and determination of methicillin resistance among coagulase negative staphylococci. Mem Inst Oswaldo Cruz. 2006;101(3):277-80.
43. Hussain Z, Stoakes L, Garrow S, Longo S, Fitzgerald V, Lanning R. Rapid detection of mecA-positive and mecA-negative coagulase-negative staphylococci by an anti-penicillin binding protein 2a slide latex agglutination test. J Clin Microbiol. 2000;38(6):2051-4.
44. Yamazumi T, Furuta I, Diekema DJ, Pfaller MA, Jones RN. Comparison of the Vitek gram-positive susceptibility 106 card, the MRSA-Screen latex agglutination test, and mecA analysis for detecting oxacillin resistance in a geographically diverse collection of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol. 2001;39(10):3633-6.
45. Louie L, Majury A, Goodfellow J, Louie M, Simor AE. Evaluation of a latex agglutination test (MRSA-Screen) for detection of oxacillin resistance in coagulase-negative Staphylococci. J Clin Microbiol. 2001;39(11):4149-51.
46. Pottumarthy S, Fritsche TR, Jones RN. Evaluation of alternative disk diffusion methods for detecting mecA-mediated oxacillin resistance in an international collection of staphylococci: validation report from the SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis. 2005;51(1):57-62.
47. Martinez F, Chandler LJ, Reisner BS, Woods GL. Evaluation of the Vitek card GPS105 and VTK-RO7.01 software for detection of oxacillin resistance in clinically relevant coagulase-negative staphylococci. J Clin Microbiol. 2001;39(10):3733-5.
48. Horstkotte MA, Knobloch JK, Rohde H, Dobinsky S, Mack D. Rapid detection of methicillin resistance in coagulase-negative Staphylococci with the VITEK 2 system. J Clin Microbiol. 2002;40(9):3291-5.
49. York MK, Gibbs L, Chehab F, Brooks GF. Comparison of PCR detection of mecA with standard susceptibility testing methods to determine methicillin resistance in coagulase-negative staphylococci. J Clin Microbiol. 1996;34(2): 249-53.
50. Kohner P, Uhl J, Kolbert C, Persing D, Cockerill F 3rd. Comparison of susceptibility testing methods with mecA gene analysis for determining oxacillin (methicillin) resistance in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus spp. J Clin Microbiol. 1999;37(9):2952-61.
51. Hackbarth CJ, Chambers HF. Methicillin-resistant Staphylococci: Detection methods and treatment of infections. Antimicrob Agents Chemother. 1989;33 (7):995-9.
52. Chambers HF. Methicillin-resistant Staphylococci: Molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10(4):781-791.
53. Schaefler S, Jones D, Perry W, Ruvinskaya L, Baradet T, Mayr E, et al. Emergence of gentamicin- and methicillin resistant Staphylococcus aureus strains in New York City hospitals. J Clin Microbiol. 1981;13:754-9.
54. Oliveira AD, Hofling-Lima AL, Belfort R Jr, Gavoso MF, Francisco W. Fluoroquinolones Susceptibilities to Methicillin Resistant and Susceptible Coagulase Negative Staphylococcus Isolated From Eye Infection. Arq Bras Oftalmol. 2007;70(2):286-9.
55. Stroman DW, Clark L, Macke L, et al. Moxifloxacin activity against quinolone resistant Staphylococcal ocular isolates [abstract]. Invest Ophthalmol Vis Sci. 2001;42(suppl 4):1377.
56. Mather R, Karenchak LM, Romanowski EG, Kowalski RP. Fourth generation fluoroquinolones: new weapons in the arsenal of ophthalmic antibiotics. Am J Ophthalmol. 2002;133(4):463-6.
57. Kowalski RP, Dhaliwal DK, Karenchak LM, Romanowski EG, Mah FS, Ritterband DC, et al. Gatifloxacin and moxifloxacin: an in vitro susceptibility comparison to levofloxacin, ciprofloxacin, and ofloxacin using bacterial keratitis isolates. Am J Ophthalmol. 2003;136(3):500-5.
58. Stroman DW, Dajcs JJ, Cupp GA, Schlech BA. In vitro and in vivo potency of moxifloxacin and moxifloxacin ophthalmic solution 0.5%, a new topical fluoroquinolone. Surv Ophthalmol. 2005;50 Suppl:S16-31.
 Endereço para correspondência:
Endereço para correspondência:
Ana Luisa Höfling-Lima
Av. Ibijaú, 331 - 17º Andar
São Paulo (SP) - CEP 04524-020
E-mail: [email protected]; [email protected]
Recebido para publicação em 16.09.2006
Última versão recebida em 10.04.2007
Aprovação em 02.05.2007
Nota Editorial: Depois de concluída a análise do artigo sob sigilo editorial e com a anuência do Dr. Paulo Ricardo de Oliveira sobre a divulgação de seu nome como revisor, agradecemos sua participação neste processo.